This example shows how the data in the package were created and saved.
Specify where to save the downloaded data. Data from a griddap() call can be verbose, so you’ll probably want a folder for it.
fil_dir <- file.path(here::here(), "inst", "extdata")
if(!dir.exists(fil_dir)) dir.create(fil_dir)Winkel Tripel projection
crs.wintri <- "+proj=wintri +lon_0=0 +lat_1=0 +x_0=0 +y_0=0 +datum=WGS84 +units=km +no_defs"
sample_raster
lats <- c(40.375, 50.375)
lons <- c(-141.875, -120.875)
df_info <- rerddap::info("ncdcOisst21Agg_LonPM180")
df <- rerddap::griddap("ncdcOisst21Agg_LonPM180", latitude = lats, longitude = lons, time = c("2021-06-19", "2021-06-19"), fields = "sst")$dataWe turn this matrix into a raster using the raster package and the rasterFromXYZ() function.
df2 <- data.frame(x=df$lon, y=df$lat, z=df$sst)
ras <- raster::rasterFromXYZ(df2, crs = "+proj=longlat")Convert to Winkel Tripel projection
ras.wintri <- raster::projectRaster(ras, crs=crs.wintri, over=TRUE)
lats.wintri <- raster::bbox(ras.wintri)[1,]
lons.wintri <- raster::bbox(ras.wintri)[2,]World coastline
world <- rnaturalearth::ne_countries(scale = "small", returnclass = "sp")
# Get rid of interior boundaries
world <- rgeos::gUnaryUnion(world)
world.wintri <- sp::spTransform(world, crs.wintri)
p <- lapply(world.wintri@polygons , slot , "Polygons")
big <- log(unlist(lapply(p[[1]], function(x) slot(x, "area")))) > 14
# Only keep the continents
trim.world.wintri <- world.wintri
trim.world.wintri@polygons[[1]]@Polygons <- trim.world.wintri@polygons[[1]]@Polygons[which(big)]
trim.world.wintri@polygons[[1]]@plotOrder <- 1:length(trim.world.wintri@polygons[[1]]@Polygons)
slot(trim.world.wintri, "polygons") <- lapply(slot(trim.world.wintri, "polygons"),
"comment<-", NULL)## Loading required package: raster## Loading required package: sp
300 km offshore line
Note, the units for world.wintri were set to km. It is important that this code is run on a polygon in a meter as opposed to longlat projection.
buffer300 <- list()
polys <- list(wintri=world.wintri, trim.wintri=trim.world.wintri)
for(j in 1:length(polys)){
p <- polys[[j]]
world.poly <- world.wintri
buff1 <- rgeos::gBuffer(world.poly, width = 300, byid = TRUE)
# erase the inner world polygon
e <- raster::erase(buff1, world.poly)
# Use the `remove.holes()` function to get only the outer line.
e300 <- spatialEco::remove.holes(spatialEco::remove.holes(e))
# Only keep the continents
e300@polygons[[1]]@Polygons <- e300@polygons[[1]]@Polygons[2]
e300@polygons[[1]]@plotOrder <- as.integer(1)
el300 <- as(e300, "SpatialLines")
df <- c()
n <- length(el300@lines[[1]]@Lines)
for (i in 1:n) {
df <- rbind(df, cbind(el300@lines[[1]]@Lines[[i]]@coords, ID = i))
}
tmp <- list(line = el300, polygon = e300, df = df, crs = crs.wintri)
buffer300[[names(polys)[j]]] <- tmp
}## Warning in sp::proj4string(x): CRS object has comment, which is lost in output
## Warning in sp::proj4string(x): CRS object has comment, which is lost in output
## Warning in sp::proj4string(x): CRS object has comment, which is lost in output
## Warning in sp::proj4string(x): CRS object has comment, which is lost in outputMake 20km coast line
Only Holes function direct adaptation from spatialEco::remove.holes()
only.holes <- function (x)
{
if (!any(which(utils::installed.packages()[, 1] %in% "maptools")))
stop("please install maptools package before running this function")
xp <- slot(x, "polygons")
holes <- lapply(xp, function(x) sapply(methods::slot(x, "Polygons"),
methods::slot, "hole"))
res <- lapply(1:length(xp), function(i) methods::slot(xp[[i]],
"Polygons")[holes[[i]]])
IDs <- row.names(x)
x.fill <- sp::SpatialPolygons(lapply(1:length(res), function(i) sp::Polygons(res[[i]],
ID = IDs[i])), proj4string = sp::CRS(sp::proj4string(x)))
methods::slot(x.fill, "polygons") <- lapply(methods::slot(x.fill,
"polygons"), maptools::checkPolygonsHoles)
methods::slot(x.fill, "polygons") <- lapply(methods::slot(x.fill,
"polygons"), "comment<-", NULL)
pids <- sapply(methods::slot(x.fill, "polygons"), function(x) methods::slot(x, "ID"))
x.fill <- sp::SpatialPolygonsDataFrame(x.fill, data.frame(row.names = pids,
ID = 1:length(pids)))
return(x.fill)
}Create buffer 280 km around the 300km line. Note because we are doing this around a line, we don’t need to erase the inner polygon as we had to do for the 300km line.
buffer20 <- list()
polys <- list(wintri=buffer300$wintri$line, trim.wintri=buffer300$trim.wintri$line)
for(j in 1:length(polys)){
p <- polys[[j]]
buff20 <- rgeos::gBuffer(el300, width=280)
# Remove the outer part of the polygons and make into lines.
e20 <- only.holes(buff20)
el20 <- as(e20, "SpatialLines")
# Convert to a dataframe also.
df <- c()
n <- length(el20@lines[[1]]@Lines)
for (i in 1:n) {
df <- rbind(df, cbind(el20@lines[[1]]@Lines[[i]]@coords, ID = i))
}
tmp <- list(line = el20, polygon = e20, df = df, crs = crs.wintri)
buffer20[[names(polys)[j]]] <- tmp
}## Warning in sp::proj4string(x): CRS object has comment, which is lost in output
## Warning in sp::proj4string(x): CRS object has comment, which is lost in outputRecreate the 300 km buffer to smooth it out a bit
Create buffer 280 km around the 300km line. Note because we are doing this around a line, we don’t need to erase the inner polygon as we had to do for the 300km line.
buff300 <- rgeos::gBuffer(el20, width=280)
e300 <- spatialEco::remove.holes(spatialEco::remove.holes(buff300))
el300 <- as(e300, "SpatialLines")
df300 <- c()
n <- length(el300@lines[[1]]@Lines)
for (i in 1:n) {
df300 <- rbind(df300, cbind(el20@lines[[1]]@Lines[[i]]@coords, ID = i))
}Coast sample locations every 100km
numOfPoints <- rgeos::gLength(el20) / 100
sample.pts.100km <- sp::spsample(el20, n = numOfPoints, type = "regular")
sample.pts.off.100km <- sp::spsample(el300, n = numOfPoints, type = "regular")
sample_points <- list(km100=sample.pts.100km, off.km100=sample.pts.off.100km)
plot(sample_raster$raster.wintri)
plot(el300, add=TRUE)
plot(el20, add=TRUE, col="red")
plot(sample.pts.100km, add=TRUE, pch=1)
plot(sample.pts.off.100km, add=TRUE)
text(sample.pts.100km@coords[,1], sample.pts.100km@coords[,2], 1:nrow(sample.pts.100km@coords), adj=-1)
text(sample.pts.off.100km@coords[,1], sample.pts.off.100km@coords[,2], 1:nrow(sample.pts.off.100km@coords), adj=1)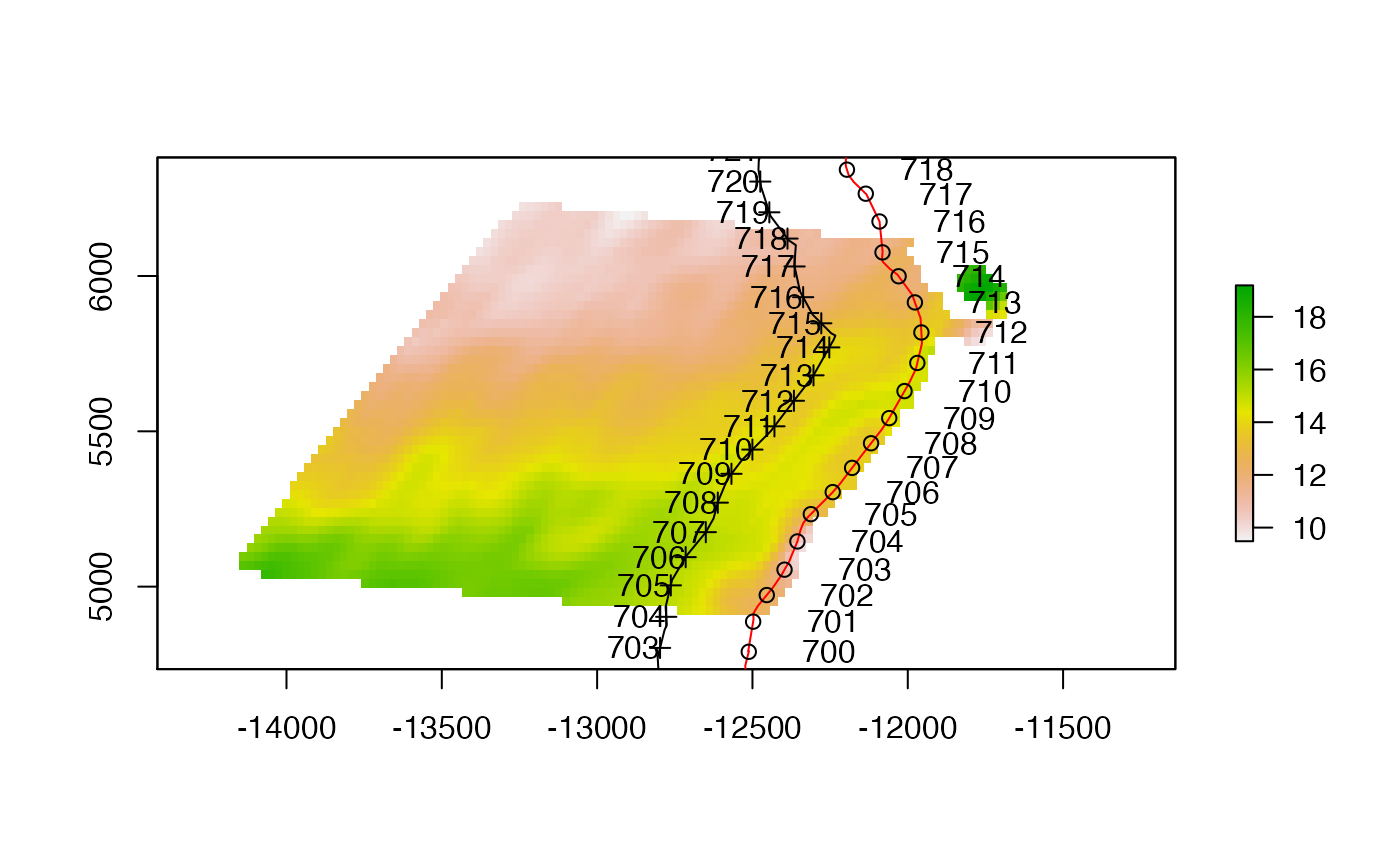
plot(e300)
bb <- raster::drawExtent()
plot(raster::crop(el300, bb))
plot(el20, add=TRUE, col="red")
plot(sample.pts.100km, add=TRUE, pch=1)
plot(sample.pts.off.100km, add=TRUE)
text(sample.pts.100km@coords[,1], sample.pts.100km@coords[,2], 1:nrow(sample.pts.100km@coords), adj=-1)
text(sample.pts.off.100km@coords[,1], sample.pts.off.100km@coords[,2], 1:nrow(sample.pts.off.100km@coords), adj=1)
plot(world.wintri, col="grey", add=TRUE)Save the objects
crs_wintri <- crs.wintri
save(crs_wintri, file = file.path(here::here(), "data/crs_wintri.rda"))
save(world, world.wintri, trim.world.wintri, file = file.path(here::here(), "data/world.rda"))
save(buffer300, buffer20, file = file.path(here::here(), "data/buffers.rda"))
save(sample_points, file = file.path(here::here(), "data/sample_points.rda"))