Gap-filling with ConvLSTM#
Author: Yifei Hang (UW Varanasi intern 2024), Jiarui Yu (UW Varanasi intern 2023)
This notebook shows how to fit a basic ConvLSTM for filling the gaps in the Chlorophyll-a data. Although you can run this tutorial on CPU, it will be much faster on GPU. We used the image quay.io/pangeo/ml-notebook:2024.08.18 for running the notebook.
import xarray as xr
import numpy as np
import dask.array as da
import matplotlib.pyplot as plt
import tensorflow as tf
from tensorflow.keras.callbacks import EarlyStopping
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import ConvLSTM2D, BatchNormalization, Conv2D, Dropout
# list all the physical devices
physical_devices = tf.config.list_physical_devices()
print("All Physical Devices:", physical_devices)
# list all the available GPUs
gpus = tf.config.list_physical_devices('GPU')
print("Available GPUs:", gpus)
# Print infomation for available GPU if there exists any
if gpus:
for gpu in gpus:
details = tf.config.experimental.get_device_details(gpu)
print("GPU Details:", details)
else:
print("No GPU available")
All Physical Devices: [PhysicalDevice(name='/physical_device:CPU:0', device_type='CPU'), PhysicalDevice(name='/physical_device:GPU:0', device_type='GPU')]
Available GPUs: [PhysicalDevice(name='/physical_device:GPU:0', device_type='GPU')]
GPU Details: {'device_name': 'NVIDIA GeForce RTX 4070 Laptop GPU', 'compute_capability': (8, 9)}
zarr_ds = xr.open_zarr(store="./参考资料/2023_INTERN_MATERIAL/INDIAN_OCEAN_025GRID_DAILY.zarr", consolidated=True) # get data
zarr_sliced = zarr_ds.sel(lat=slice(35, -5), lon=slice(45,90)) # choose long and lat
all_nan_CHL = np.isnan(zarr_sliced.CHL).all(dim=["lon", "lat"]).compute() # find sample indices where CHL is NaN
zarr_CHL = zarr_sliced.sel(time=(all_nan_CHL == False)) # select samples with CHL not NaN
zarr_CHL = zarr_CHL.sortby('time')
zarr_CHL = zarr_CHL.sel(time=slice('2020-01-01', '2020-12-31'))
zarr_CHL
<xarray.Dataset> Size: 711MB
Dimensions: (time: 366, lat: 149, lon: 181)
Coordinates:
* lat (lat) float32 596B 32.0 31.75 31.5 ... -4.5 -4.75 -5.0
* lon (lon) float32 724B 45.0 45.25 45.5 ... 89.5 89.75 90.0
* time (time) datetime64[ns] 3kB 2020-01-01 ... 2020-12-31
Data variables: (12/19)
CHL (time, lat, lon) float32 39MB dask.array<chunksize=(25, 149, 181), meta=np.ndarray>
CHL_uncertainty (time, lat, lon) float32 39MB dask.array<chunksize=(25, 149, 181), meta=np.ndarray>
adt (time, lat, lon) float32 39MB dask.array<chunksize=(25, 149, 181), meta=np.ndarray>
air_temp (time, lat, lon) float32 39MB dask.array<chunksize=(25, 149, 181), meta=np.ndarray>
curr_dir (time, lat, lon) float32 39MB dask.array<chunksize=(25, 149, 181), meta=np.ndarray>
curr_speed (time, lat, lon) float32 39MB dask.array<chunksize=(25, 149, 181), meta=np.ndarray>
... ...
ug_curr (time, lat, lon) float32 39MB dask.array<chunksize=(25, 149, 181), meta=np.ndarray>
v_curr (time, lat, lon) float32 39MB dask.array<chunksize=(25, 149, 181), meta=np.ndarray>
v_wind (time, lat, lon) float32 39MB dask.array<chunksize=(25, 149, 181), meta=np.ndarray>
vg_curr (time, lat, lon) float32 39MB dask.array<chunksize=(25, 149, 181), meta=np.ndarray>
wind_dir (time, lat, lon) float32 39MB dask.array<chunksize=(25, 149, 181), meta=np.ndarray>
wind_speed (time, lat, lon) float32 39MB dask.array<chunksize=(25, 149, 181), meta=np.ndarray>
Attributes: (12/17)
creator_email: minhphan@uw.edu
creator_name: Minh Phan
creator_type: person
date_created: 2023-07-19
geospatial_lat_max: 32.0
geospatial_lat_min: -12.0
... ...
geospatial_lon_units: degrees_east
source: Earth & Space Research (ESR), Copernicus Clim...
summary: Daily mean of 0.25 x 0.25 degrees gridded dat...
time_coverage_end: 2022-12-31T23:59:59
time_coverage_start: 1979-01-01T00:00:00
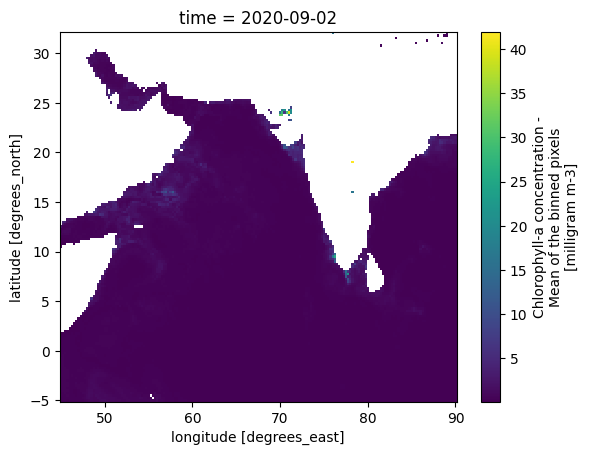
title: Climate Data for Coastal Upwelling Machine Le...p = zarr_CHL.sel(time='2020-09-02').CHL.plot(y='lat', x='lon')
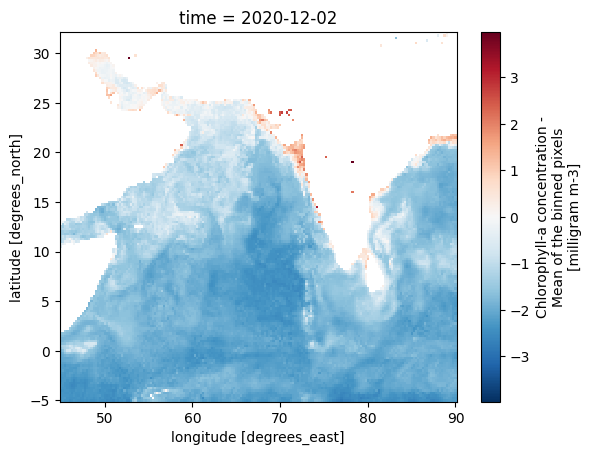
np.log(zarr_CHL.sel(time='2020-12-02').CHL).plot(y='lat', x='lon')
<matplotlib.collections.QuadMesh at 0x23ca9d2fa90>
def log_label(data, label):
data_logged = data.copy()
data_logged[label] = np.log(data[label]).copy()
return data_logged
# Add more preprocessing later
def preprocess_data(data, features, label):
# log CHL
data_logged = log_label(data, label)
# save all features and labels in one list
sel_data_list = []
for var in (features + [label]):
sel_var_data = data_logged[var]
sel_var_data = da.where(da.isnan(sel_var_data), 0.0, sel_var_data)
sel_data_list.append(sel_var_data)
# reassign datatype of list to dask array
sel_data_da = da.array(sel_data_list)
# of dimensions (var, time, lat, lon)
return sel_data_da
#
def data_prep(data, window_size=5):
X_orig = data
y_orig = data[-1]
X_orig = np.moveaxis(X_orig, 0, -1)
length = X_orig.shape[0]
X, y = [], []
for i in range(length - window_size):
X.append(X_orig[i: i + window_size])
y.append(y_orig[i + window_size])
X, y = da.array(X), da.array(y)
return X, y
def time_series_split(X, y, split_ratio):
total_length = X.shape[0]
train_end = int(total_length * split_ratio[0])
val_end = int(total_length * (split_ratio[0] + split_ratio[1]))
X_train, y_train = X[:train_end], y[:train_end]
X_val, y_val = X[train_end: val_end], y[train_end: val_end]
X_test, y_test = X[val_end:], y[val_end:]
return (X_train, y_train,
X_val, y_val,
X_test, y_test)
features = ['sst', 'so'] # Curr Features: Sea Surface Temp (K), Sea Salinity Concentration (m**-3 or PSL). [Excluding Topography/Bathymetry (m)]
label = 'CHL' # Curr Label: : chlorophyll-a concentration (mg/m**3) [Not taking uncertainty into consideration for now]
model_data = preprocess_data(zarr_CHL, features, label)
window_size = 5
X, y = data_prep(model_data, window_size=window_size)
split_ratio = [.7, .2, .1]
X_train, y_train, X_val, y_val, X_test, y_test = time_series_split(X, y, split_ratio)
y_train.shape
(252, 149, 181)
def create_model_ConvLSTM(input_shape=(5, 149, 181, 3)):
model = Sequential()
model.add(ConvLSTM2D(filters=32, kernel_size=(3, 3),
input_shape=input_shape,
padding='same', return_sequences=False))
model.add(BatchNormalization())
model.add(Dropout(0.2))
model.add(Conv2D(filters=64,
kernel_size=(3, 3),
padding='same',
activation='relu'
))
model.add(BatchNormalization())
model.add(Dropout(0.2))
model.add(Conv2D(filters=32,
kernel_size=(3, 3),
padding='same',
activation='relu'
))
model.add(BatchNormalization())
model.add(Dropout(0.2))
model.add(Conv2D(filters=1,
kernel_size=(3, 3),
padding='same',
activation='linear'
))
return model
model = create_model_ConvLSTM()
model.summary()
Model: "sequential"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
conv_lstm2d (ConvLSTM2D) (None, 149, 181, 32) 40448
batch_normalization (BatchN (None, 149, 181, 32) 128
ormalization)
dropout (Dropout) (None, 149, 181, 32) 0
conv2d (Conv2D) (None, 149, 181, 64) 18496
batch_normalization_1 (Batc (None, 149, 181, 64) 256
hNormalization)
dropout_1 (Dropout) (None, 149, 181, 64) 0
conv2d_1 (Conv2D) (None, 149, 181, 32) 18464
batch_normalization_2 (Batc (None, 149, 181, 32) 128
hNormalization)
dropout_2 (Dropout) (None, 149, 181, 32) 0
conv2d_2 (Conv2D) (None, 149, 181, 1) 289
=================================================================
Total params: 78,209
Trainable params: 77,953
Non-trainable params: 256
_________________________________________________________________
model.compile(optimizer='adam', loss='mae', metrics=['mae'])
early_stop = EarlyStopping(patience=10, restore_best_weights=True)
train_dataset = tf.data.Dataset.from_tensor_slices((X_train, y_train))
train_dataset = train_dataset.shuffle(buffer_size=1024).batch(8)
val_dataset = tf.data.Dataset.from_tensor_slices((X_val, y_val))
val_dataset = val_dataset.batch(8)
history = model.fit(train_dataset, epochs=50, validation_data=val_dataset, callbacks=[early_stop])
Epoch 1/50
32/32 [==============================] - 13s 260ms/step - loss: 1.0611 - mae: 1.0611 - val_loss: 0.7293 - val_mae: 0.7293
Epoch 2/50
32/32 [==============================] - 8s 236ms/step - loss: 0.5555 - mae: 0.5555 - val_loss: 0.6904 - val_mae: 0.6904
Epoch 3/50
32/32 [==============================] - 8s 237ms/step - loss: 0.3589 - mae: 0.3589 - val_loss: 0.5267 - val_mae: 0.5267
Epoch 4/50
32/32 [==============================] - 8s 235ms/step - loss: 0.3039 - mae: 0.3039 - val_loss: 0.4278 - val_mae: 0.4278
Epoch 5/50
32/32 [==============================] - 8s 244ms/step - loss: 0.2711 - mae: 0.2711 - val_loss: 0.3880 - val_mae: 0.3880
Epoch 6/50
32/32 [==============================] - 8s 236ms/step - loss: 0.2492 - mae: 0.2492 - val_loss: 0.3104 - val_mae: 0.3104
Epoch 7/50
32/32 [==============================] - 8s 239ms/step - loss: 0.2358 - mae: 0.2358 - val_loss: 0.2944 - val_mae: 0.2944
Epoch 8/50
32/32 [==============================] - 8s 235ms/step - loss: 0.2261 - mae: 0.2261 - val_loss: 0.2752 - val_mae: 0.2752
Epoch 9/50
32/32 [==============================] - 8s 234ms/step - loss: 0.2110 - mae: 0.2110 - val_loss: 0.2692 - val_mae: 0.2692
Epoch 10/50
32/32 [==============================] - 8s 235ms/step - loss: 0.2070 - mae: 0.2070 - val_loss: 0.2399 - val_mae: 0.2399
Epoch 11/50
32/32 [==============================] - 8s 235ms/step - loss: 0.2002 - mae: 0.2002 - val_loss: 0.2560 - val_mae: 0.2560
Epoch 12/50
32/32 [==============================] - 8s 240ms/step - loss: 0.2038 - mae: 0.2038 - val_loss: 0.2250 - val_mae: 0.2250
Epoch 13/50
32/32 [==============================] - 8s 237ms/step - loss: 0.2039 - mae: 0.2039 - val_loss: 0.4885 - val_mae: 0.4885
Epoch 14/50
32/32 [==============================] - 8s 242ms/step - loss: 0.2034 - mae: 0.2034 - val_loss: 0.1741 - val_mae: 0.1741
Epoch 15/50
32/32 [==============================] - 8s 240ms/step - loss: 0.1953 - mae: 0.1953 - val_loss: 0.2007 - val_mae: 0.2007
Epoch 16/50
32/32 [==============================] - 8s 246ms/step - loss: 0.1901 - mae: 0.1901 - val_loss: 0.1622 - val_mae: 0.1622
Epoch 17/50
32/32 [==============================] - 8s 240ms/step - loss: 0.1837 - mae: 0.1837 - val_loss: 0.1780 - val_mae: 0.1780
Epoch 18/50
32/32 [==============================] - 8s 241ms/step - loss: 0.1822 - mae: 0.1822 - val_loss: 0.3413 - val_mae: 0.3413
Epoch 19/50
32/32 [==============================] - 8s 242ms/step - loss: 0.1771 - mae: 0.1771 - val_loss: 0.1499 - val_mae: 0.1499
Epoch 20/50
32/32 [==============================] - 8s 243ms/step - loss: 0.1877 - mae: 0.1877 - val_loss: 0.4606 - val_mae: 0.4606
Epoch 21/50
32/32 [==============================] - 8s 242ms/step - loss: 0.1908 - mae: 0.1908 - val_loss: 0.1608 - val_mae: 0.1608
Epoch 22/50
32/32 [==============================] - 8s 237ms/step - loss: 0.1743 - mae: 0.1743 - val_loss: 0.2386 - val_mae: 0.2386
Epoch 23/50
32/32 [==============================] - 8s 238ms/step - loss: 0.1712 - mae: 0.1712 - val_loss: 0.1379 - val_mae: 0.1379
Epoch 24/50
32/32 [==============================] - 8s 237ms/step - loss: 0.1735 - mae: 0.1735 - val_loss: 0.2289 - val_mae: 0.2289
Epoch 25/50
32/32 [==============================] - 8s 235ms/step - loss: 0.1718 - mae: 0.1718 - val_loss: 0.1371 - val_mae: 0.1371
Epoch 26/50
32/32 [==============================] - 8s 236ms/step - loss: 0.1714 - mae: 0.1714 - val_loss: 0.1574 - val_mae: 0.1574
Epoch 27/50
32/32 [==============================] - 8s 236ms/step - loss: 0.1760 - mae: 0.1760 - val_loss: 0.3918 - val_mae: 0.3918
Epoch 28/50
32/32 [==============================] - 8s 236ms/step - loss: 0.1724 - mae: 0.1724 - val_loss: 0.2902 - val_mae: 0.2902
Epoch 29/50
32/32 [==============================] - 8s 236ms/step - loss: 0.1796 - mae: 0.1796 - val_loss: 0.1230 - val_mae: 0.1230
Epoch 30/50
32/32 [==============================] - 8s 237ms/step - loss: 0.1686 - mae: 0.1686 - val_loss: 0.2143 - val_mae: 0.2143
Epoch 31/50
32/32 [==============================] - 8s 238ms/step - loss: 0.1635 - mae: 0.1635 - val_loss: 0.1600 - val_mae: 0.1600
Epoch 32/50
32/32 [==============================] - 8s 234ms/step - loss: 0.1619 - mae: 0.1619 - val_loss: 0.1123 - val_mae: 0.1123
Epoch 33/50
32/32 [==============================] - 8s 237ms/step - loss: 0.1618 - mae: 0.1618 - val_loss: 0.1696 - val_mae: 0.1696
Epoch 34/50
32/32 [==============================] - 8s 238ms/step - loss: 0.1656 - mae: 0.1656 - val_loss: 0.2791 - val_mae: 0.2791
Epoch 35/50
32/32 [==============================] - 8s 237ms/step - loss: 0.1713 - mae: 0.1713 - val_loss: 0.1272 - val_mae: 0.1272
Epoch 36/50
32/32 [==============================] - 8s 241ms/step - loss: 0.1715 - mae: 0.1715 - val_loss: 0.1695 - val_mae: 0.1695
Epoch 37/50
32/32 [==============================] - 8s 237ms/step - loss: 0.1664 - mae: 0.1664 - val_loss: 0.2333 - val_mae: 0.2333
Epoch 38/50
32/32 [==============================] - 8s 237ms/step - loss: 0.1717 - mae: 0.1717 - val_loss: 0.2303 - val_mae: 0.2303
Epoch 39/50
32/32 [==============================] - 8s 236ms/step - loss: 0.1606 - mae: 0.1606 - val_loss: 0.1642 - val_mae: 0.1642
Epoch 40/50
32/32 [==============================] - 8s 236ms/step - loss: 0.1570 - mae: 0.1570 - val_loss: 0.1952 - val_mae: 0.1952
Epoch 41/50
32/32 [==============================] - 8s 236ms/step - loss: 0.1646 - mae: 0.1646 - val_loss: 0.1192 - val_mae: 0.1192
Epoch 42/50
32/32 [==============================] - 8s 238ms/step - loss: 0.1621 - mae: 0.1621 - val_loss: 0.1814 - val_mae: 0.1814
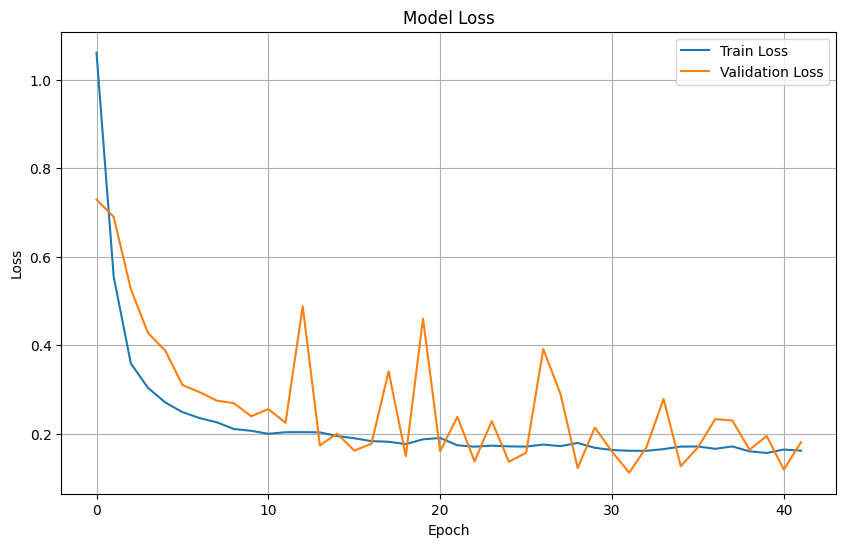
# Plot training & validation loss values
plt.figure(figsize=(10, 6))
plt.plot(history.history['loss'], label='Train Loss')
plt.plot(history.history['val_loss'], label='Validation Loss')
plt.title('Model Loss')
plt.xlabel('Epoch')
plt.ylabel('Loss')
plt.legend(loc='upper right')
plt.grid(True)
plt.show()
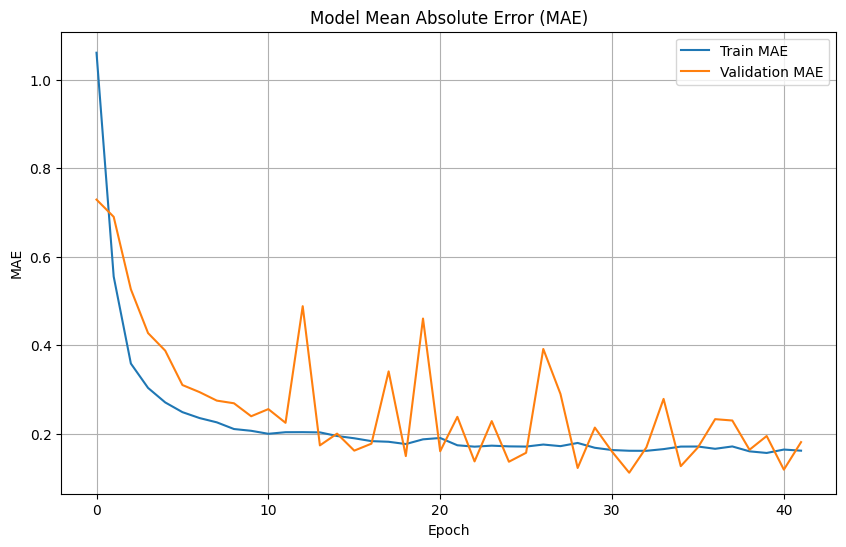
# Plot training & validation MAE values
plt.figure(figsize=(10, 6))
plt.plot(history.history['mae'], label='Train MAE')
plt.plot(history.history['val_mae'], label='Validation MAE')
plt.title('Model Mean Absolute Error (MAE)')
plt.xlabel('Epoch')
plt.ylabel('MAE')
plt.legend(loc='upper right')
plt.grid(True)
plt.show()
# Prepare test dataset
test_dataset = tf.data.Dataset.from_tensor_slices((X_test, y_test))
test_dataset = test_dataset.batch(4)
# Evaluate the model on the test dataset
test_loss, test_mae = model.evaluate(test_dataset)
print(f"Test Loss: {test_loss}")
print(f"Test MAE: {test_mae}")
10/10 [==============================] - 1s 56ms/step - loss: 0.1045 - mae: 0.1045
Test Loss: 0.10454108566045761
Test MAE: 0.1045411005616188
X
|
||||||||||||||||
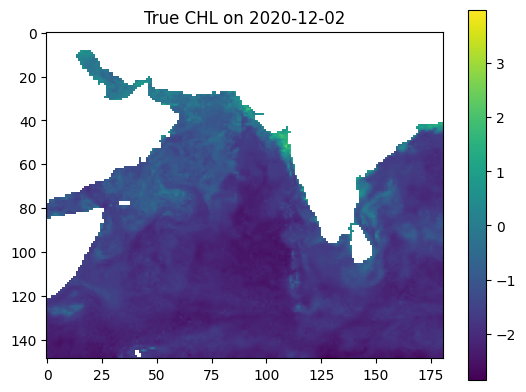
date_to_predict = '2020-12-02'
true_output = np.log(zarr_CHL.sel(time=date_to_predict).CHL)
date_index = (np.datetime64(date_to_predict) - np.datetime64('2020-01-01')).item().days
input = X[date_index - window_size]
input = np.array(input)
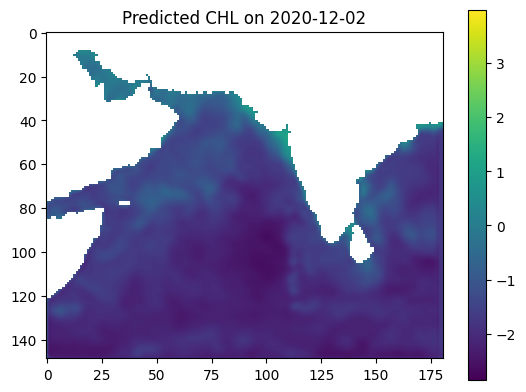
predicted_output = model.predict(input[np.newaxis, ...])[0]
predicted_output = predicted_output[:,:,0]
land_mask = np.load(r"E:\24SU Varanasi Intern\参考资料\2023_Intern_Material\land_mask_nc.npy")
predicted_output[land_mask] = np.nan
# true_output_2 = (model_data[-1])[date_index]
vmax = np.nanmax((true_output, predicted_output))
vmin = np.nanmin((true_output, predicted_output))
plt.imshow(true_output, vmin=vmin, vmax=vmax)
plt.colorbar()
plt.title(f'True CHL on {date_to_predict}')
plt.show()
plt.imshow(predicted_output, vmin=vmin, vmax=vmax)
plt.colorbar()
plt.title(f'Predicted CHL on {date_to_predict}')
plt.show()
1/1 [==============================] - 0s 10ms/step
def compute_mae(y_true, y_pred):
mask = ~np.isnan(y_true) & ~np.isnan(y_pred)
return np.mean(np.abs(y_true[mask] - y_pred[mask]))
predicted_mae = compute_mae(np.array(true_output), predicted_output)
print(f"MAE between Predicted Output and True Output: {predicted_mae}")
prev_day_dt64 = np.datetime64(date_to_predict) - np.timedelta64(1, 'D')
prev_day = np.datetime_as_string(prev_day_dt64, unit='D')
prev_day_CHL = np.log(zarr_CHL.sel(time=prev_day).CHL)
last_frame_mae = compute_mae(np.array(prev_day_CHL), np.array(true_output))
print(f"MAE between Last Input Frame and True Output: {last_frame_mae}")
MAE between Predicted Output and True Output: 0.16125544905662537
MAE between Last Input Frame and True Output: 0.13173514604568481
input.shape
input[-1,:,:,-1].shape
(149, 181)
# t = numpy.datetime64('2012-06-30T20:00:00.000000000-0400')
# numpy.datetime_as_string(t, unit='D')
last_day = np.datetime64(date_to_predict) - np.timedelta64(1, 'D')
last_day_str = np.datetime_as_string(last_day, unit='D')
last_day_str
'2020-09-01'
# (np.datetime64('2020-01-03') - np.datetime64('2020-01-01')) / np.timedelta64(1, 'D')
(np.datetime64('2020-01-03') - np.datetime64('2020-01-01')).item().days
2
